About us
About BOARDS database
Welcome to BOARDS database, the Blanket Overarching
Antimicrobial-Resistance gene
Database with Structural investigation.
This database allows researchers to easily search for information on various antibiotic
resistance(AMR) genes.
BOARDS database provides genetic and structural information of various AMR genes.
Using this tool, microbiologists can easily explore and search for information about the multiple antibiotic resistance (AMR) genes identified in an organism. Currently, BOARDS DB provides an AMR dashboard, database statistics, the information search tables on all AMR Genes in the database, and frequently occurring SNP information. In particular, BOARDS DB provides structural information of all AMR genes (even SNP structures) included as well.
How to Use this site
Begin by selecting a tab from the navigation bar on the start page. The AMR dashboard page covers the distribution of AMR genes in pathogens based on the results analyzed through the BOARDS DB targeting 8 types of antibiotic-resistant bacteria, including ESKAPE pathogens. Additionally, from the Download tab at the bottom of the navigation bar, you can download all AMR gene data from the BOARDS DB and conduct your own custom analysis.
The BOARDS DB provides a plain text file of all AMR gene data recorded in "Fasta" (for nucleotide sequence) or "FAA" (for amino-acid sequence) file format. You can also download predicted protein structure information and perform structural biological custom analysis.
Once you've reached the AMR dashboard, two pieces of plotting data will be shown. You can browse and investigate the response chart showing the increase in Whole-Genome Sequencing (WGS) data of the major monitored strains and the ESBL pan-resistome identified within the corresponding pathogen. For more information on the webpage menu component, see the section below.
It is our hope that BOARDS DB will enrich research by providing easy access to this compendium of AMR gene datasets and structural information of the corresponding gene product. If you would like to perform a broad analysis of the AMR gene, it is expected that the BOARDS DB will be able to provide detailed information relevant to your research.
BOARDS DB Statistics
This page visualizes multiple statistical figures for BOARDS DB and plots them so that you can understand the current status of Database in details.
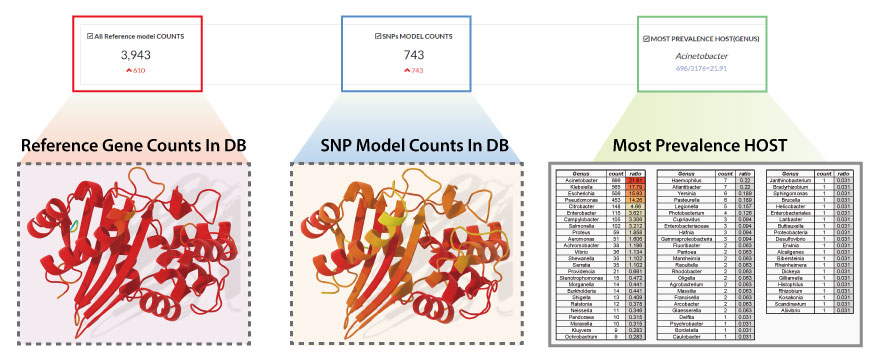
Statistics: Top Panel
- The Leftmost Information: The leftmost information in the panel indicates the number of AMR genes included in the DB, and can be called a "reference model" included in the BOARDS DB.
- The Center Information: The information located in the center represents the number of SNP models included in the BOARDS DB.
- The Rightmost Information: The information located on the far right re-expresses the origin of the antibiotic resistance gene based on the metadata of the gene included in the BOARDS DB, and it expresses that the most antibiotic resistance gene was detected in the organism.
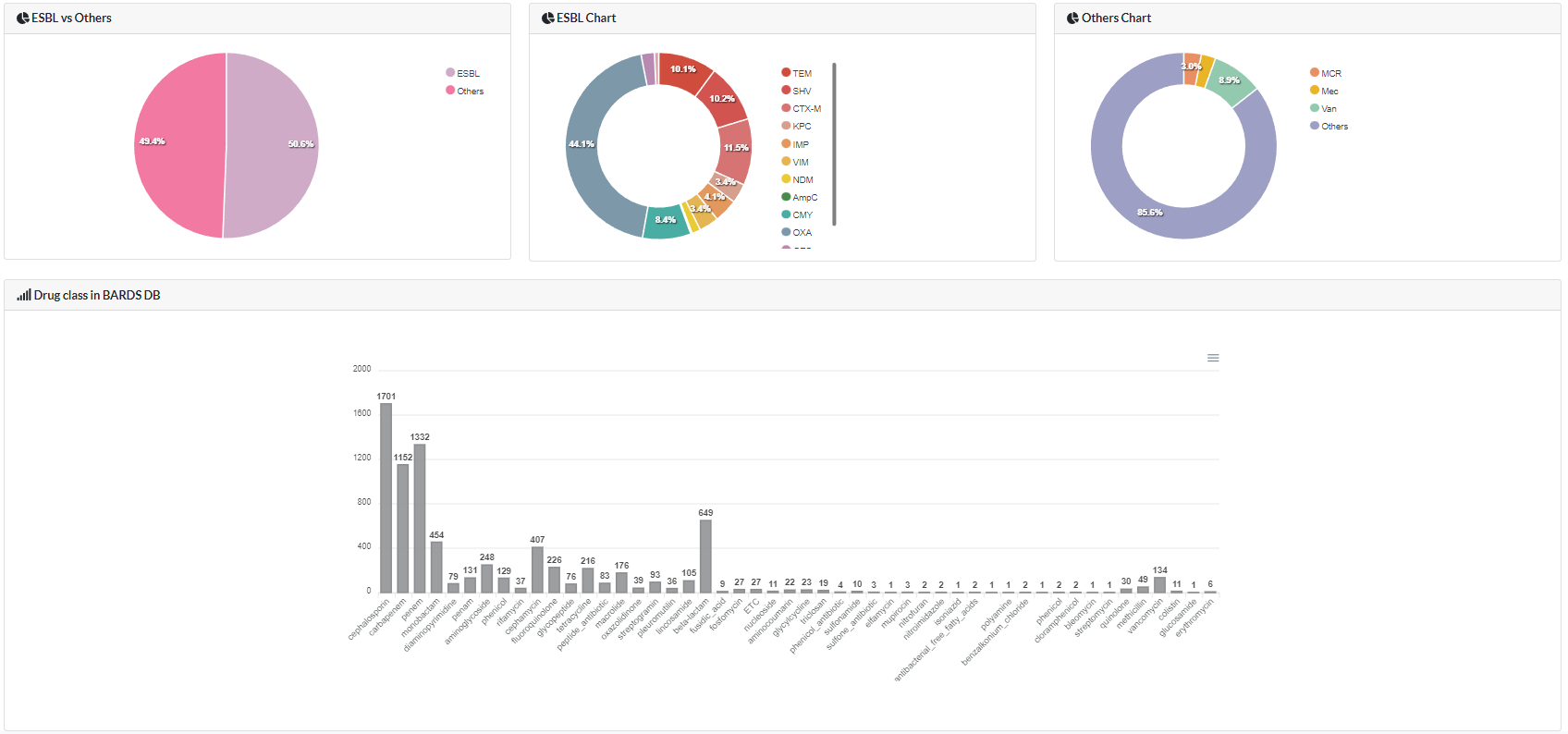
Statistics: Middle Panel
The middle panel shows the results of categorization and plotting based on the
metadata of genes contained in the BOARDS DB.
As is already known, ESBL is a key gene to be monitored as it is spreading very
rapidly. Therefore, the BOARDS DB plotted the ESBL genes in great detail.
In addition, the bar plot located at the bottom expresses the current status of
drug resistance classes of all AMR genes listed in the BOARDS DB.
BOARDS Table
This page tabulates all AMR genes listed by the BOARDS DB.
The table displayed on this page indicates the following information.
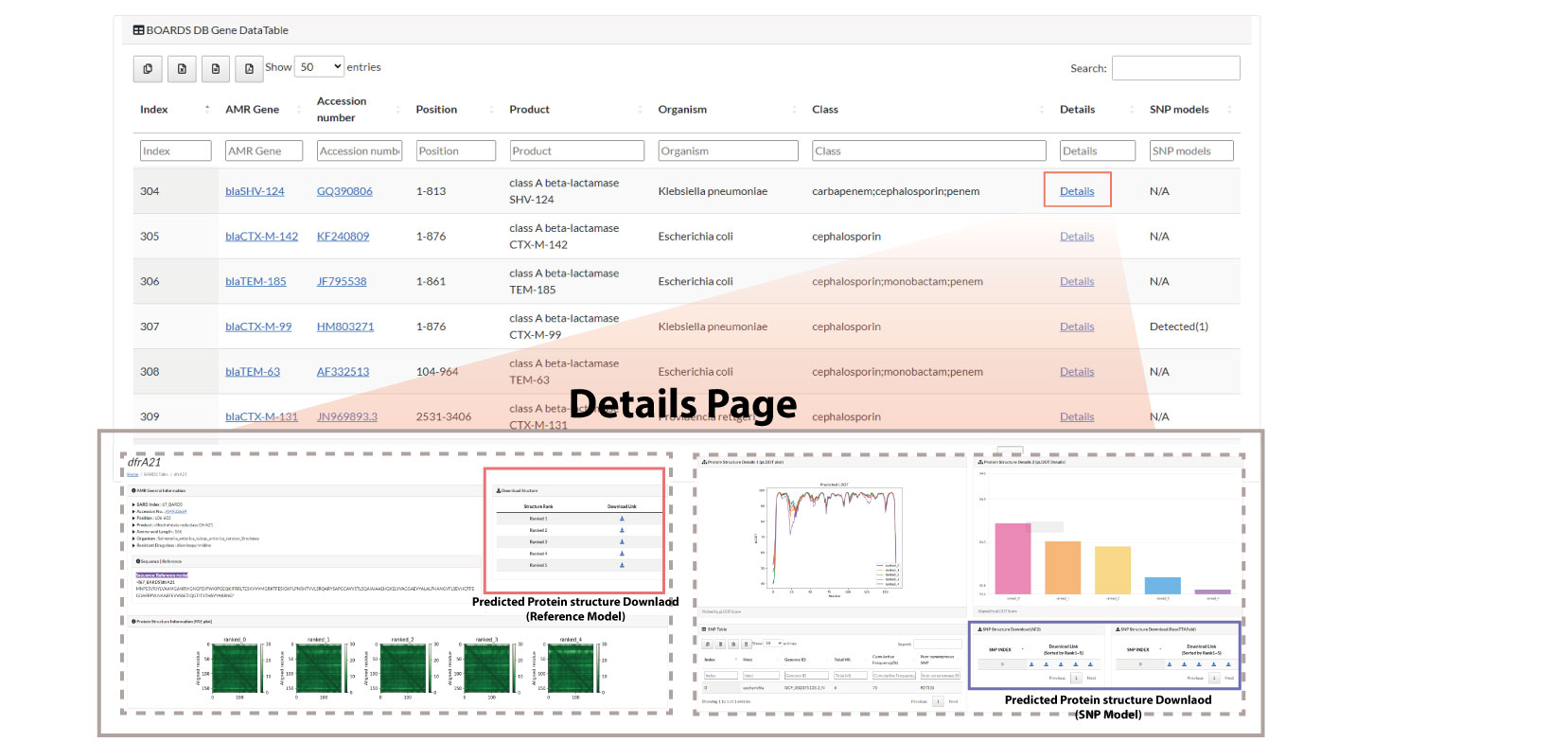
- Index: Displays the index of BOARDS db.
- AMR Gene: Displays the name of the AMR Gene.
- Accession number: Indicates the NCBI accession number of AMR Gene. If you press the corresponding accession number, you will be connected to NCBI.
- Position: Indicates the position of the corresponding AMR gene in the genome.
- Organism: Displays the organism information of the corresponding AMR gene.
-
Class: Displays drug resistance information of the corresponding AMR gene. Indicate
the relationship conferring or contributing to antibiotic resistance by the AMR gene.
Much of the information was collected by the CARD database, and only insufficient information was secured through literature review. -
Details: Provides a hyperlink to a page that will display structural information for
the corresponding AMR gene. You will be directed to a page that provides structural
information about the reference model. Also, if there are SNP models that occur frequently
in that gene, it can be covered within the hyperlinks provided. The Details page provides
the following items.
- AMR General information: Shows general information of AMR gene.
- Reference model Sequence: Displays sequence information of the corresponding AMR gene (Reference model).
- Protein Structure Information (PAE plot): Shows a PAE (Predicted Aligned Error) plot of the predicted protein structure calculated by AlphaFold2 using the sequence information of the corresponding AMR gene (reference model). For a detailed definition of PAE plot, see the page on AlphaFold2 page.
- Protein Structure Information (pLDDT plot): Displays the pLDDT plot of the predicted protein structure of the corresponding AMR gene (Reference model). For a definition of pLDDT, see AlphaFold2's page.
- Protein Structure Information (Average pLDDT): The pLDDT score of the predicted protein structure is expressed as a bar plot. Detailed score information is displayed by hovering the mouse over it.
- SNP Table: If there is a frequently occurring SNP model within the corresponding AMR gene (Reference model), it is displayed as a table. If there is no SNP model, the message "No data available in table" appears.
- Download:
- Download StructureYou can download the predicted protein structure information of the Reference model. There are a total of 5 predicted protein structure information, and the Top 5 sorted by pLDDT score are provided.
- SNP Structure Download (AF2): Activated only when there is a SNP model. The download link provides the structurally predicted model with AlphaFold2. If the SNP model exists, 5 download links of the model indexed based on the SNP Table are provided.
- SNP Structure Download (RoseTTAFold): Activated only when there is a SNP model. The download link provides a model for structural prediction with RoseTTAFold. If the SNP model exists, 5 download links of the model indexed based on the SNP Table are provided.